Genomic Exploration of Functional Plant
Ginseng Genome Analysis
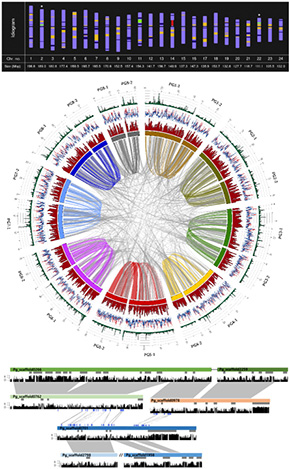
Our laboratory was the first in the world to complete and report the genome sequencing of ginseng, a representative medicinal crop of Korea. The ginseng genome contains approximately 3.6 gigabase pairs (Gbp) of genetic information distributed across 24 chromosome pairs, with about 90% of its sequence composed of repetitive elements. Additionally, ginseng exhibits a highly complex genome structure characteristic of an allotetraploid, which originated from the hybridization of different species. This genomic complexity contributes to enhanced environmental adaptability, such as the ability to overwinter, distinguishing it from its diploid relatives. By studying such intricate genomes, we can uncover the fascinating secrets of plant biology.
Transposable element
Numerous studies have demonstrated that transposable elements (TEs) are not only responsible for genome size variation in crops but also play a crucial role in diverse phenotypic traits. Our laboratory has previously identified that a major transposable element in ginseng, the PgDel LTR retrotransposon, has significantly contributed to species distribution and evolution. Currently, we are conducting comprehensive research on transposable elements in ginseng and Araliaceae plants, including their genome-wide identification, evolutionary mechanisms, and regulatory functions. Furthermore, we aim to establish a database of transposable elements associated with ginseng and Araliaceae species.
Discovery of Functional Plant Resources and Molecular Breeding
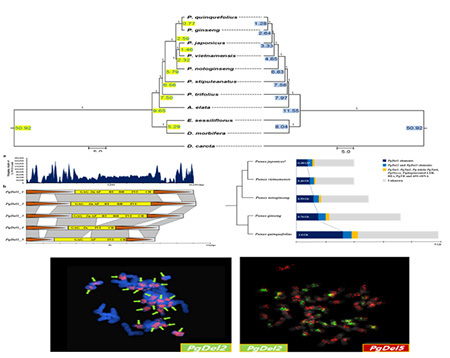
Establishment of a Molecular Breeding System for Ginseng
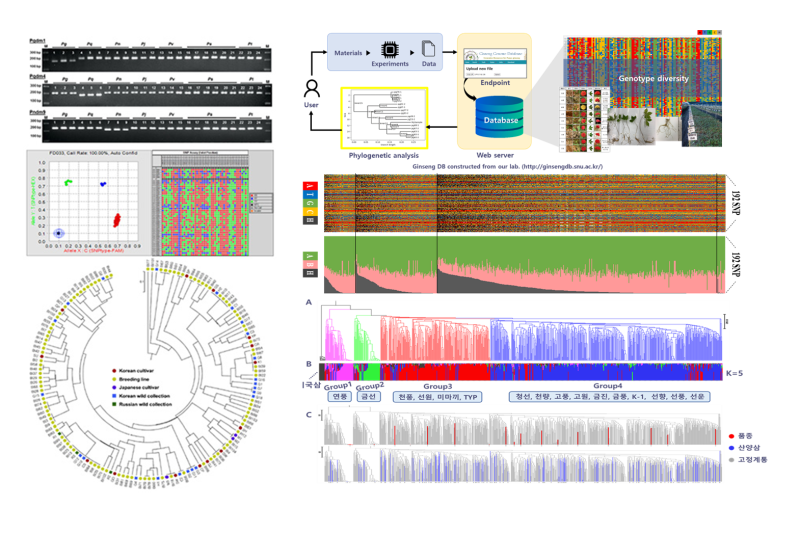
Ginseng requires a four-year growth period to produce seeds, making breeding a time-intensive and laborious process. Our laboratory has completed the whole-genome sequencing of ginseng and is leveraging this data to design an advanced molecular breeding system. We have applied DNA molecular markers to over 1,400 ginseng accessions of different varieties and geographic origins, constructing a comprehensive ginseng genetic database. This database facilitates the assessment of genetic diversity and fixation within ginseng landraces and cultivars, thereby enabling the rapid and precise breeding of high-quality ginseng varieties.
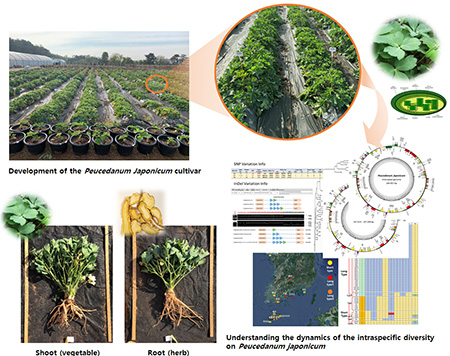
Breeding and Molecular Breeding System for Sikbangpung
Our laboratory is actively engaged in developing new cultivars of Sikbangpung(Peucedanum japonicum) with superior traits, such as vegetable-specific and medicinal-use-specific characteristics. We are selecting elite Sikbangpung individuals and advancing generations through selfing techniques. Additionally, to support molecular breeding, we are conducting genomic studies to explore intraspecific genetic diversity within Sikbangpung.
DNA Barcoding and Evolutionary Studies
DNA barcoding
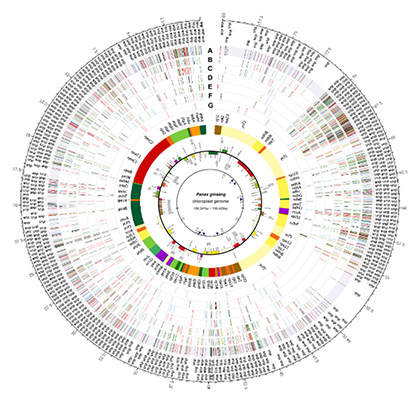
DNA barcoding is a technique used to identify species by analyzing their unique DNA sequences. For plant species identification, chloroplast genomes and nuclear 45S ribosomal DNA are commonly utilized. While early studies relied on short DNA fragments, recent advancements have introduced the super barcode concept, which employs entire chloroplast genome sequences for species identification. Our laboratory has developed a novel method—de novo assembly using low-coverage whole-genome sequencing (dnaLCW)—which enables the simultaneous analysis of chloroplast genomes and nuclear 45S ribosomal DNA using minimal sequencing data. To date, we have applied this technique to over 400 plant species, contributing to genome analysis and species identification.
The genomic variations identified from these completed genome sequences are utilized to develop species- and cultivar-specific DNA markers. These markers facilitate the authentication of raw plant materials and commercialized herbal products, ensuring a transparent medicinal plant distribution system. Furthermore, we are actively conducting genome-based evolutionary research, including phylogenetic relationship establishment, evolutionary lineage analysis, and divergence time estimation among plant species.
Metabolite Biosynthesis and Enhancement
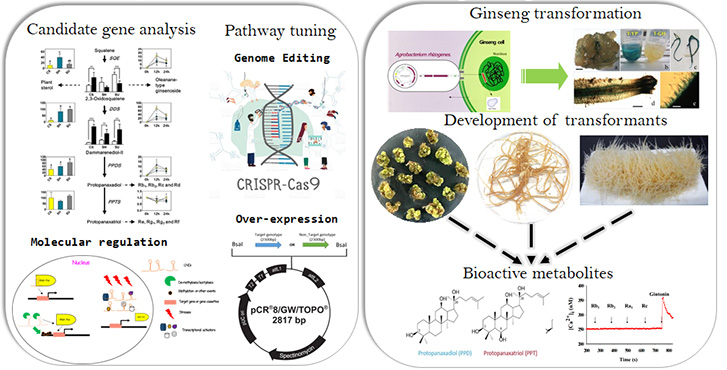
Functional Characterization of Ginseng Genes and Enhancement of Ginsenoside Content
Our laboratory employs integrative omics technologies to refine the biosynthetic pathway of ginsenosides by identifying and validating novel genes and UDP-glycosyltransferases (UGTs). We are actively working on enhancing ginsenoside content through genome editing techniques, as well as gene overexpression in ginseng callus and adventitious roots.
